|
Plenary
Lecture
Artificial Restriction DNA Cutter for
Manipulation of Huge DNA

Professor Makoto Komiyama
Research Center for Advanced Science and Technology
University of Tokyo, Tokyo
153-8904 Japan
E-mail:
komiyama@mkomi.rcast.u-tokyo.ac.jp
Abstract:
Current molecular biology is based on site-selective
scission of DNA by restriction enzymes. However,
site-specificity of naturally occurring restriction
enzymes is too low to manipulate huge DNA. In order to
solve this problem, we developed man-made tools which
cut double-stranded DNA at desired site (artificial
restriction DNA cutter; ARCUT).1) These tools are
composed of (1) Ce(IV)/EDTA complex as molecular
scissors and (2) two pseudo-complementary PNAs (blue
lines in Fig. 1). By changing the sequences and the
lengths of PNA strands, the scission-site and
site-specificity are freely chosen.
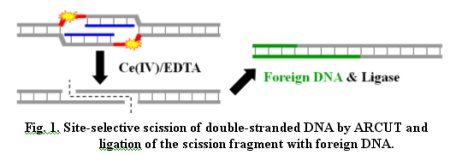
With the use
of ARCUT, even huge DNAs (e.g., the whole genome of
human beings) were selectively cut at target site. The
resultant scission fragments were ligated with foreign
DNAs (green lines in Fig. 1) and expressed in mammalian
cells. Applications of ARCUT for promotion of homologous
recombination in human cells are also presented.
Brief Biography of the Speaker:
Makoto Komiyama graduated from the University of Tokyo
in 1970, and got his Ph.D. from the same University in
1975. After spending four years at Northwestern
University (USA) as a postdoctoral fellow, he became an
assistant professor at the University of Tokyo, and then
an associate professor at University of Tsukuba. From
1991, he has been a professor of the University of
Tokyo. His main research area is bioorganic and
bioinorganic chemistry. He received Awards for Young
Scientist from the Chemical Society of Japan, Japan IBM
Science Award, Award from the Rare Earth Society of
Japan, Inoue Prize for Science, The Award of the Society
of Polymer Science, Japan, Award from Cyclodextrin
Society of Japan, and others.
|

